Import Different Types of File in Python for Analysis - Part 1
Typically, data scientists will work with enormous types of file. Having the knowledge of these types of file and importing them properly will speed up the process of data analyzing. In this post I will show how to import various types of data for anylyzing purpose using Python.
Firstly, I am importing some of the Python modules I will be using throughout the tutorial as follows
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
Working with flat files
In this part I will import Flat files such as txt, csv and so on.
The following code block will open a text file with read mode and then I am reading the file using read() method read the file. Finally I am printing the file.
# opening
file = open('data/seaslug.txt', mode='r')
# read a file
myFile = file.read()
print(myFile)
Time Percent
99 0.067
99 0.133
99 0.067
99 0
Previously we read the whole file at once. We can even read file line by line as well. The readline() method do the job for us easily.
# importing text files line by line
with open('data/seaslug.txt') as file:
print(file.readline())
print(file.readline())
Time Percent
99 0.067
We can use numpy to import data as well. Here I am importing csv file using numpy. In numpy loadtext() function we are passing two arguments - the file and the delimter which is ‘,’ for csv file.
# importing data with numpy
# Import package
import numpy as np
# Assign filename to variable: file
file = 'data/mnist_kaggle_some_rows.csv'
# Load file as array: digits
digits = np.loadtxt(file, delimiter=',')
# Select and reshape a row
im = digits[9, 1:]
im_sq = np.reshape(im, (28, 28))
# Plot reshaped data (matplotlib.pyplot already loaded as plt)
plt.imshow(im_sq, interpolation='nearest')
plt.show()
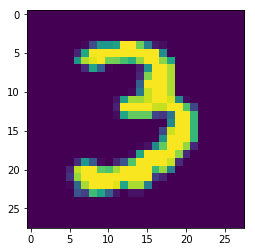
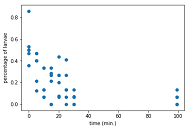
Sometimes we need to import data without column name or title. We can do so easily by one more additional argument into numpy loadtxt() function called skiprows=1. It will skip the first row of the dataset.
# reading file without the name of the column-header
file = 'data/seaslug.txt'
data = np.loadtxt(file, delimiter='\t', skiprows=1)
# printing 5 rows to make sure first row (column name) is not there
print(data[:5])
[[9.90e+01 6.70e-02]
[9.90e+01 1.33e-01]
[9.90e+01 6.70e-02]
[9.90e+01 0.00e+00]
[9.90e+01 0.00e+00]]
plt.scatter(data[:, 0], data[:, 1])
plt.xlabel('time (min.)')
plt.ylabel('percentage of larvae')
plt.show()
We can import file which has both numeric and text value in it. By using numpy recfromcsv() function this job can be done very easily.
# importing file using recfromcsv funcion
file = 'data/titanic_sub.csv'
data = np.recfromcsv(file)
print(data[:3])
[(1, 0, 3, b'male', 22., 1, 0, b'A/5 21171', 7.25 , b'', b'S')
(2, 1, 1, b'female', 38., 1, 0, b'PC 17599', 71.2833, b'C85', b'C')
(3, 1, 3, b'female', 26., 0, 0, b'STON/O2. 3101282', 7.925 , b'', b'S')]
Pandas is the best way to import file which gives a lot of flexibility. We can import csv file by using Pandas read_csv() function. And then we can look into the first few line (by default 10) of data by using pandas head method
# Assign filename: file
file = 'data/titanic_sub.csv'
# Import file: data
data = pd.read_csv(file, sep=',', comment='#', na_values='NaN')
# Print the head of the DataFrame
print(data.head())
PassengerId Survived Pclass Sex Age SibSp Parch \
0 1 0 3 male 22.0 1 0
1 2 1 1 female 38.0 1 0
2 3 1 3 female 26.0 0 0
3 4 1 1 female 35.0 1 0
4 5 0 3 male 35.0 0 0
Ticket Fare Cabin Embarked
0 A/5 21171 7.2500 NaN S
1 PC 17599 71.2833 C85 C
2 STON/O2. 3101282 7.9250 NaN S
3 113803 53.1000 C123 S
4 373450 8.0500 NaN S
Working with spreadsheets
We can import excel spreadsheet file by using pandas ExcelFile() funtion. In the following lines of code, first we are loading the spreadsheet by using ExcelFile() function. And then we can use sheet_names() to know no of sheets in the spreadsheet. In the following file it has two sheets names ‘2004’ and ‘2002’. I am loading this sheets in padas dataframe usnig parse() function.
# importing xlsx
file = 'data/battledeath.xlsx'
# load spreadsheet
data = pd.ExcelFile(file)
# printing sheetnames
print('Sheet Names: \n',data.sheet_names, '\n')
# Load a sheet into a DataFrame by name: df1
df1 = data.parse('2004')
# Print the head of the DataFrame df1
print(df1.head())
print('\n')
# Load a sheet into a DataFrame by index: df2
df2 = data.parse('2002')
# Print the head of the DataFrame df2
print(df2.head())
Sheet Names:
['2002', '2004']
War(country) 2004
0 Afghanistan 9.451028
1 Albania 0.130354
2 Algeria 3.407277
3 Andorra 0.000000
4 Angola 2.597931
War, age-adjusted mortality due to 2002
0 Afghanistan 36.083990
1 Albania 0.128908
2 Algeria 18.314120
3 Andorra 0.000000
4 Angola 18.964560
Working with SAS file
We can import sas file by using sas7bdat module. Here I am getting the file using SAS7BDAF and saving it as data frame with to_data_frame() method.
# Import sas7bdat package
from sas7bdat import SAS7BDAT
# Save file to a DataFrame: df_sas
with SAS7BDAT('data/sales.sas7bdat') as file:
df_sas = file.to_data_frame()
# Print head of DataFrame
print(df_sas.head())
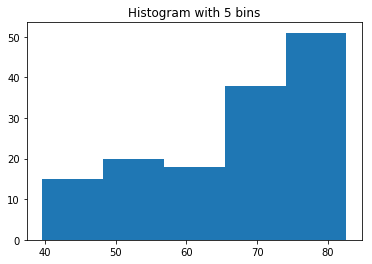
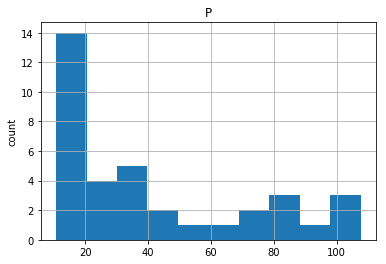
# Plot histogram of DataFrame features (pandas and pyplot already imported)
pd.DataFrame.hist(df_sas[['P']])
plt.ylabel('count')
plt.show()
YEAR P S
0 1950.0 12.9 181.899994
1 1951.0 11.9 245.000000
2 1952.0 10.7 250.199997
3 1953.0 11.3 265.899994
4 1954.0 11.2 248.500000
Working with STATA file
We can easily import stata file with pandas read_stata function. You can learn about stata from wikipedia. Below I am have one state dataset called disarea. I am importing this dataset using pandas read_stata funciton and printing 3 rows of data by using pandas head() method.
# loading stata file in pandas dataframe
file = 'data/disarea.dta'
df = pd.read_stata(file)
# printing first 3 lines
print(df.head(3))
wbcode country disa1 disa2 disa3 disa4 disa5 disa6 disa7 disa8 \
0 AFG Afghanistan 0.00 0.00 0.76 0.73 0.0 0.0 0.00 0.0
1 AGO Angola 0.32 0.02 0.56 0.00 0.0 0.0 0.56 0.0
2 ALB Albania 0.00 0.00 0.02 0.00 0.0 0.0 0.00 0.0
... disa16 disa17 disa18 disa19 disa20 disa21 disa22 disa23 \
0 ... 0.0 0.0 0.0 0.00 0.0 0.0 0.00 0.02
1 ... 0.0 0.4 0.0 0.61 0.0 0.0 0.99 0.98
2 ... 0.0 0.0 0.0 0.00 0.0 0.0 0.00 0.00
disa24 disa25
0 0.00 0.00
1 0.61 0.00
2 0.00 0.16
[3 rows x 27 columns]
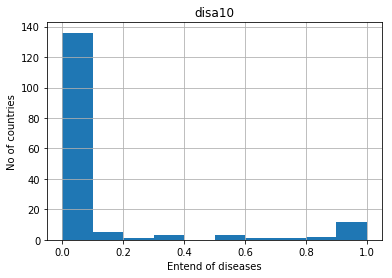
# ploting histogram
pd.DataFrame.hist(df[['disa10']])
plt.xlabel("Entend of diseases")
plt.ylabel("No of countries")
Text(0,0.5,'No of countries')
Working with hdf5 file
# Working with hdf5 file
import h5py
# assign filename to file
file = 'data/L-L1_LOSC_4_V1-1126259446-32.hdf5'
#load file: data
data = h5py.File(file, 'r')
# printing the keys of the file
for key in data.keys():
print(key)
meta
quality
strain
# get the hdf5 group: group
group = data['strain']
# check out of keys of group
for key in group.keys():
print(key)
# set variable equal to time series data: strain
strain = data['strain']['Strain'].value
# num of time points to sample
num_samples = 10000
# set the vector
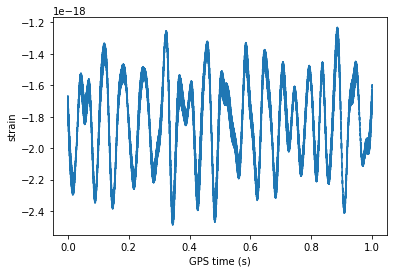
time = np.arange(0, 1 , 1/num_samples)
# ploting data
plt.plot(time, strain[:num_samples])
plt.xlabel('GPS time (s)')
plt.ylabel('strain')
plt.show()
Strain
Working with Matlab file
Another popular type of file is called matlab file with extension .mat at the end. We can import such file by using scipy.io module in Python. Here I am loading the file using scipy.io.loadmat() funcion by passing the mat file. And then checking the file type. As we can see it belongs to Python dict class. That means we extract the keys out of it. We can go furthe deep by digging the value of these keys.
# Importing matlab file
import scipy.io
# assigning filename to file
file = 'data/ja_data2.mat'
# loading matlab file
mat = scipy.io.loadmat(file)
# check type of the file
print('Type of mat file', type(mat), '\n')
# extracting keys from mat file
for key in mat.keys():
print(key)
# Print the type of the value corresponding to the key 'CYratioCyt'
print(type(mat['CYratioCyt']))
# Print the shape of the value corresponding to the key 'CYratioCyt'
print(np.shape(mat['CYratioCyt']))
Type of mat file <class 'dict'>
__header__
__version__
__globals__
rfpCyt
rfpNuc
cfpNuc
cfpCyt
yfpNuc
yfpCyt
CYratioCyt
<class 'numpy.ndarray'>
(200, 137)
# Print the type of the value corresponding to the key 'CYratioCyt'
print(type(mat['CYratioCyt']))
# Print the shape of the value corresponding to the key 'CYratioCyt'
print(np.shape(mat['CYratioCyt']))
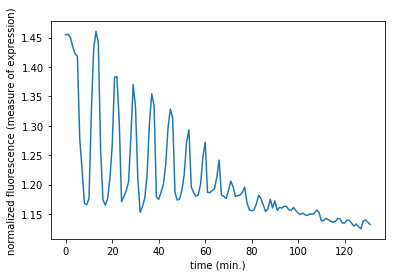
# Subset the array and plot it
data = mat['CYratioCyt'][25, 5:]
fig = plt.figure()
plt.plot(data)
plt.xlabel('time (min.)')
plt.ylabel('normalized fluorescence (measure of expression)')
plt.show()
<class 'numpy.ndarray'>
(200, 137)
Working with Database file
It’s quite obvious to work with databases while working in company. That’s why good understanding of databases is the key skill of data scientist. We can use sqlalchemy to work with databases.
In the following lines of code first I am importing create_engine from sqlalchemy which I will be using to establish connection with databases. I am creating the engine by passing the appropriate argument such as the type of file working with is sqlite. As we know, databases are consist of tables. We can extrct the table by using table_names() method which will return a list of tables names avaailable in the database file.
# Working with databases
# importing necessary module
from sqlalchemy import create_engine
# Create engine
engine = create_engine('sqlite:///data/Chinook.sqlite')
# creating a list with table names contains in engine
table_names = engine.table_names()
# printing table names
print(table_names)
['Album', 'Artist', 'Customer', 'Employee', 'Genre', 'Invoice', 'InvoiceLine', 'MediaType', 'Playlist', 'PlaylistTrack', 'Track']
Now we know what tables are avaiable in our datadase file. It’s time to execute some sqlite queries. First we establish connection using connect() method and then execute any query with the execute() funtion. Below I am seleting everything from Album table and passing it to the pandas DataFrame() funciton to create dataframe for me.
# opening engine connection
con = engine.connect()
# Perform query
rs = con.execute('SELECT * FROM Album')
# saving to dataframe
df = pd.DataFrame(rs.fetchall(), columns=rs.keys())
# close connection
con.close()
# printing our datafrmae
print(df.head())
AlbumId Title ArtistId
0 1 For Those About To Rock We Salute You 1
1 2 Balls to the Wall 2
2 3 Restless and Wild 2
3 4 Let There Be Rock 1
4 5 Big Ones 3
Below is the shorter version to establish connection and execute queries on database where we don’t need to care about closing the connection. Below codeblock pretty does the same as above except here I am executing several queries and capturing into different dataframes.
# better way
with engine.connect() as con:
rs = con.execute('SELECT LastName, Title FROM Employee')
df = pd.DataFrame(rs.fetchmany(size=10), columns = rs.keys())
rs2 = con.execute('SELECT * FROM Employee ORDER BY BirthDate')
df2 = pd.DataFrame(rs2.fetchall(), columns=rs2.keys())
rs3 = con.execute("SELECT Title, Name FROM Album INNER JOIN Artist on Album.ArtistID = Artist.ArtistID")
df3 = pd.DataFrame(rs3.fetchall(), columns=rs3.keys())
print(df.head())
print('\n')
print(df2.head(2),'\n')
print(df3.head(3))
LastName Title
0 Adams General Manager
1 Edwards Sales Manager
2 Peacock Sales Support Agent
3 Park Sales Support Agent
4 Johnson Sales Support Agent
EmployeeId LastName FirstName Title ReportsTo \
0 4 Park Margaret Sales Support Agent 2.0
1 2 Edwards Nancy Sales Manager 1.0
BirthDate HireDate Address City State \
0 1947-09-19 00:00:00 2003-05-03 00:00:00 683 10 Street SW Calgary AB
1 1958-12-08 00:00:00 2002-05-01 00:00:00 825 8 Ave SW Calgary AB
Country PostalCode Phone Fax \
0 Canada T2P 5G3 +1 (403) 263-4423 +1 (403) 263-4289
1 Canada T2P 2T3 +1 (403) 262-3443 +1 (403) 262-3322
Email
0 margaret@chinookcorp.com
1 nancy@chinookcorp.com
Title Name
0 For Those About To Rock We Salute You AC/DC
1 Balls to the Wall Accept
2 Restless and Wild Accept
Here is the Pandas way of interacting with database file. We can use pandas read_sql_query() function do our job by passing the query we want to execute and the engine. [engine is defined eartier]
df = pd.read_sql_query('SELECT * FROM Employee WHERE EmployeeId >=6 ORDER BY BirthDate', engine)
# Print head of DataFrame
print(df.head())
EmployeeId LastName FirstName Title ReportsTo BirthDate \
0 8 Callahan Laura IT Staff 6 1968-01-09 00:00:00
1 7 King Robert IT Staff 6 1970-05-29 00:00:00
2 6 Mitchell Michael IT Manager 1 1973-07-01 00:00:00
HireDate Address City State Country \
0 2004-03-04 00:00:00 923 7 ST NW Lethbridge AB Canada
1 2004-01-02 00:00:00 590 Columbia Boulevard West Lethbridge AB Canada
2 2003-10-17 00:00:00 5827 Bowness Road NW Calgary AB Canada
PostalCode Phone Fax Email
0 T1H 1Y8 +1 (403) 467-3351 +1 (403) 467-8772 laura@chinookcorp.com
1 T1K 5N8 +1 (403) 456-9986 +1 (403) 456-8485 robert@chinookcorp.com
2 T3B 0C5 +1 (403) 246-9887 +1 (403) 246-9899 michael@chinookcorp.com
I have posted this post in medium.